- Find a Doctor
-
For Parents
- Before Your Visit
- During Your Visit
- After Your Visit
- More Resources for Parents
Patient & Visitor Resources -
Services
- Locations
-
About Us
- About Childrens
- Find it Fast
- Additional Resources
Find it FastAdditional Resources - MyCHP
ALERT:
There is construction in and around UPMC Children’s Hospital that is affecting the traffic flow – please allow for extra time traveling into the hospital.
- Find a Doctor
- For Parents
-
Services
-
Frequently Searched Services
- Asthma Center
- Brain Care Institute (Neurology & Neurosurgery)
- Cancer
- UPMC Children's Express Care
- Ear, Nose, & Throat (ENT)
- Emergency Medicine
- Endocrinology
- Gastroenterology
- Heart Institute
- Genetic & Genomic Medicine
- Infectious Diseases
- Nephrology
- Newborn Medicine
- Primary Care
- Pulmonary Medicine
- Rheumatology
- Surgery
- Transplant Programs
- See All Services
-
Frequently Searched Services
- Locations
- About Us
- MyCHP
- I Want To
- More Links








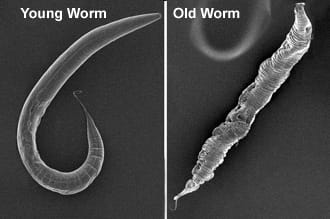 We study longevity genes in Caenorhabditis elegans, a model organism with a short lifespan of about three weeks. C. elegans displays many anatomical and functional changes that accompany human aging, as seen here in the scanning electron microscopy images of young and old C. elegans from our lab. Worm genes show strong homology with their human counterparts. Indeed, the most well known longevity pathway, the insulin/IGF1 signaling (IIS) cascade, was first identified for its role in aging in worms. Since then, many fundamental discoveries in the biology of aging have been made in C. elegans, and it continues to be a remarkably valuable system for aging research.
We study longevity genes in Caenorhabditis elegans, a model organism with a short lifespan of about three weeks. C. elegans displays many anatomical and functional changes that accompany human aging, as seen here in the scanning electron microscopy images of young and old C. elegans from our lab. Worm genes show strong homology with their human counterparts. Indeed, the most well known longevity pathway, the insulin/IGF1 signaling (IIS) cascade, was first identified for its role in aging in worms. Since then, many fundamental discoveries in the biology of aging have been made in C. elegans, and it continues to be a remarkably valuable system for aging research.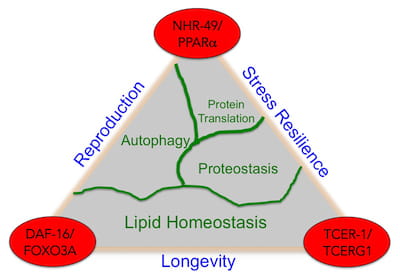 While it is well recognized that increasing age impairs reproductive fitness, how the germline of an animal influences aging of its somatic cells is not known. Recent discoveries in worms, flies and mice have shown that signals from reproductive tissues indeed influence the length and quality of life of the whole organism.
While it is well recognized that increasing age impairs reproductive fitness, how the germline of an animal influences aging of its somatic cells is not known. Recent discoveries in worms, flies and mice have shown that signals from reproductive tissues indeed influence the length and quality of life of the whole organism.
 Proteasomal Regulation of Aging
Proteasomal Regulation of Aging






